2025
254
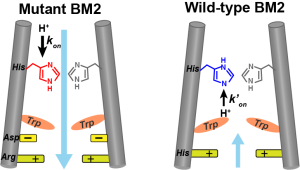
Y. Pankratova and M. Hong, “Sidechain Structures of the Proton-Selective Histidine and Gating Tryptophan in Influenza BM2 Reveal Both Conservation and Variation of the Proton Conduction Mechanism”, Biochemistry revised (2025).
253
A.J. Dregni and M. Hong, “Impact of Lysine Acetylation Mimetics on the Structure of Full-Length Tau Fibrils”, ACS Chem. Neuroscience revised (2025).
252
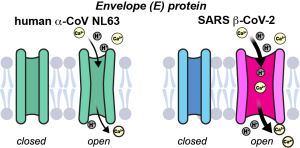
I. Sucec, B.Q. Xia, N.H. Somberg, Y. Wang, H. Jo, S. Li, B. Perrone, Z. Gao and M. Hong, “Ion Channel Structure and Function of the MERS Coronavirus E Protein”, Sci. Advances in press (2025).
251
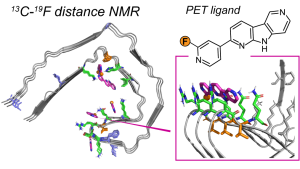
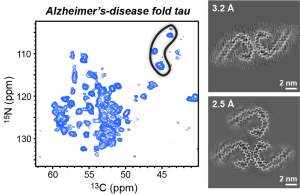
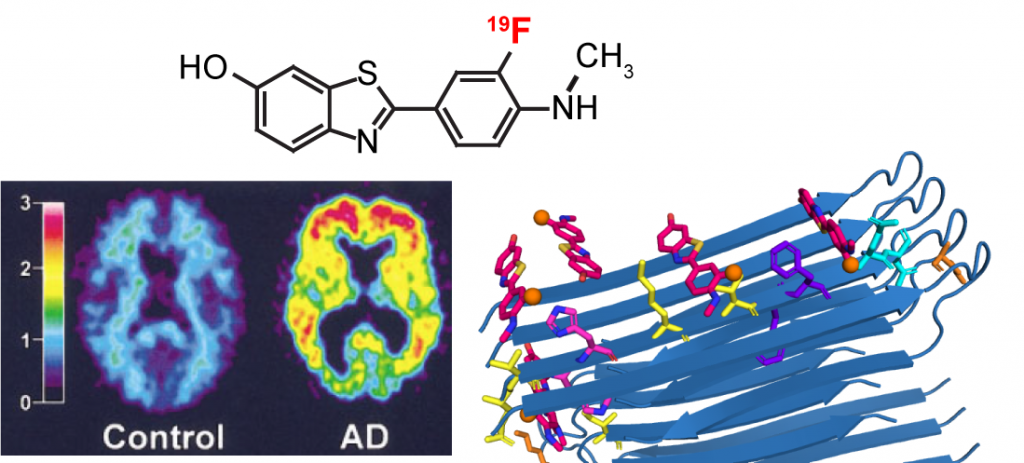
F.N. Angehrn, P. Duan, J.Y. Zhang, and M. Hong, “ Binding Sites of a PET Ligand in Tau Fibrils with the Alzheimer’s Disease Fold From 19F NMR ”, Biochemistry 64, 1624-1635 (2025).
250
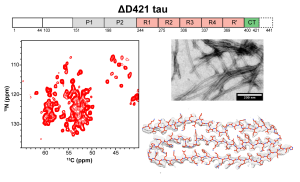
N. El Mammeri, P. Duan, and M. Hong, “ Structures of ΔD421 Truncated Tau Fibrils”, J. Mol. Biol. 437, 169051 (2025).
248
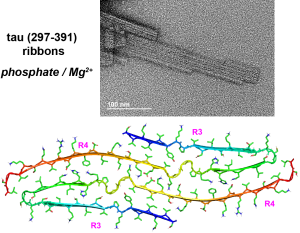
N. El Mammeri, P. Duan, and M. Hong, “Pseudo-Phosphorylated Tau Forms Paired Helical Filaments in the Presence of High-Curvature Cholesterol-Containing Lipid Membranes”, J. Am. Chem. Soc. 147, 2510-2522 (2025).
2024
247
J. Medeiros-Silva, Y. Pankratova, I. Sucec, A.J. Dregni, and M. Hong, “Polar Networks Mediate Ion Conduction of the SARS-CoV-2 Envelope Protein”, J. Am. Chem. Soc. 147, 746-757 (2025).
246
P. Duan, A. J. Dregni, H. Xu, V. M. Y. Lee, and M. Hong, “Alzheimer’s Disease Seeded Tau Forms Paired Helical Filaments Yet Lacks Seeding Potential”, J. Biol. Chem. 300, 107730 (2024).
245
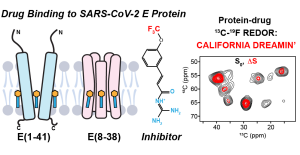
N.H. Somberg, I. Sucec, J. Medeiros-Silva, H. Jo, R. Beresis, A. M. Syed, J. A. Doudna and M. Hong, “Oligomeric State and Drug Binding of the SARS-CoV-2 E Transmembrane Domain are Sensitive to the Ectodomain”, J. Am. Chem. Soc. , 146, 24537-24552 (2024).
244
Y. Pankratova, M.J. McKay, C. Ma, H. Tan, J. Wang, and M. Hong, “Structure and Dynamics of the Proton-Selective Histidine and the Gating Tryptophan in an Inward Rectifying Hybrid Influenza B and A Virus M2 Proton Channel“, Phys. Chem. Chem. Phys., 26, 20629-20644 (2024).
243
P. Duan, N. El Mammeri and M. Hong, “Milligram-Scale Assembly and NMR Fingerprint of Tau Fibrils Adopting the Alzheimer’s Disease Fold”, J. Biol. Chem. 107326 (2024).
242
D. Tryfona, Y. Pankratova, P. Duan, M. Hong, and P. Dupree, “Altering the substitution and cross-linking of glucuronoarabinoxylans affects cell wall architecture in Brachypodium distachyon“, New Phytologist, 242, 524-543 (2024).
241
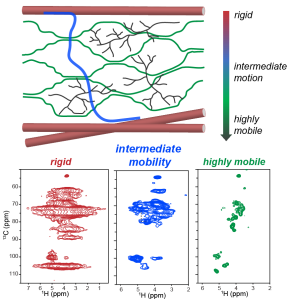
P. Duan and M. Hong, “Selective Detection of Intermediate-Amplitude Motion by Solid-State NMR“, J. Phys. Chem., 128, 2293−2303 (2024).
240
I. Sucec, Y. Pankratova, M. Parasar, and M. Hong, “Transmembrane Conformation of the Envelope Protein of an Alpha Human Coronavirus, NL63“, Protein Science, 33, e4923 (2024).
239
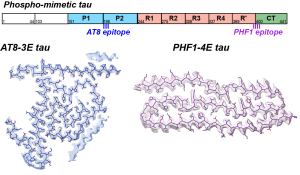
N. El Mammeri$, A.J. Dregni$, P. Duan and M. Hong, “Structures of AT8 and PHF1 Phospho-Mimetic Tau: Insights into the Posttranslational Modification Code of Tau Aggregation“, Proc. Natl. Acad. Sci. U.S.A, 121, e2316175121 (2024).
238
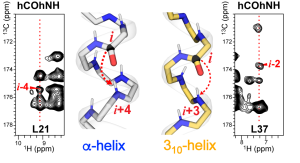
J. Medeiros-Silva and M. Hong, “Distinguishing Different Hydrogen-Bonded Helices in Proteins by Efficient 1H-Detected Three-Dimensional Solid-State NMR “, Biochemistry, 63, 181-190 (2024).
2023
237
Duan, A.J. Dregni, N. El Mammeri and M. Hong, “Structure of the Non-Helical Filament of the Alzheimer’s Disease Tau Core “, Proc. Natl. Acad. Sci. U.S.A. 120, e2310067120 (2023).
236
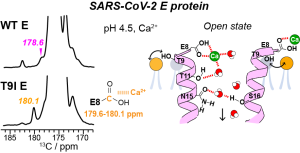
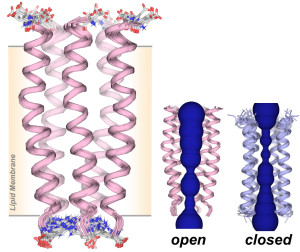
J. Medeiros-Silva, A. J. Dregni, N. H. Somberg, and M. Hong, “Atomic Structure of the Open SARS-CoV-2 E Viroporin“, Sci. Advances, 9, eadi9007 (2023).
235
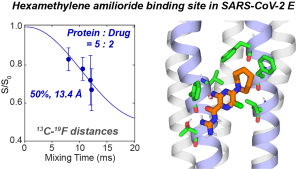
N. H. Somberg, J. Medeiros-Silva, H. Jo, J. Wang, W. F. DeGrado, and M. Hong, “Hexamethylene Amiloride Binds the SARS-CoV-2 Envelope Protein at the Protein-Lipid Interface “, Protein Science, e4755 (2023).
234
I. Sučec, N. El Mammeri, A.J. Dregni, and M. Hong, “Rapid Determination of the Topology of Oligomeric α‑Helical Membrane Proteins by Water- and Lipid-Edited Methyl NMR“, J. Phys. Chem. B, 127, 7518-7530 (2023).
233
N. El Mammeri, P. Duan, A.J. Dregni, and M. Hong, “Amyloid Fibril Structures of Tau: Conformational Plasticity of the Second Microtubule-Binding Repeat“, Sci. Adv., 9, eadh4731 (2023)
232
N. El Mammeri, O. Gampp, P. Duan and M. Hong, “Membrane-Induced Tau Amyloid Fibrils“, Commun. Biol. 6, 467 (2023).
231
A. A. Shcherbakov, M. Brousseau, K. A. Henzler-Wildman, and M. Hong, “Microsecond Motion of the Bacterial Transporter EmrE in Lipid Bilayers“, J. Am. Chem. Soc. 145, 10104-10115 (2023).
230
T. Kratochvil, L.C. Watkins, M. Mravic, N.H. Somberg, J.L. Thomaston, J. M. Nicoludis, Lijun Liu, M. Hong, G. A. Voth, W.F. DeGrado, “Transient Water Wires Mediate Selective Proton Transport in Designed Channel Proteins“. Nature Chem. 15, 1012-1021 (2023).
229
A. J. Dregni, M.J. McKay, W. Surya, M. Queralt-Martin, J. Medeiros-Silva, H. K. Wang, V. Aguilella, J. Torres, M. Hong*, The Cytoplasmic Domain of the SARS-CoV-2 Envelope Protein Assembles into a b-Sheet Bundle in Lipid Bilayers, J. Mol. Biol. 435, 167966 (2023).
2022
228
S. Kim, M. Lee, M. Hong, and N. Holten-Andersen, “Quantitative Correlation between Bound Water and Mechanical Stress-relaxation in Dehydrated Metal-Coordinate Polymer Networks“, Chem. Materials, 34, 10329-10337 (2022).
227
N.H. Somberg, W. W. Wu, J. Medeiros-Silva, H. Jo, W. F. DeGrado, and M. Hong “The SARS-CoV-2 Envelope Protein Forms Pentamers in Lipid Bilayers“, Biochemistry, 61, 2280-2294 (2022).
226
P. Duan, A.J. Dregni, and M. Hong, “Solid-State NMR 19F-1H-15N Correlation Experiments for Resonance Assignment and Distance Measurements of Multi-fluorinated Proteins“, J. Phys. Chem. 126, 7021-7032 (2022).
225
N. El-Mammeri, A.J. Dregni, P. Duan, H. K. Wang, and M. Hong, “Microtubule-binding core of the tau protein“, Sci. Adv. 8, eabo4459 (2022)
224
A. J. Dregni$, P. Duan$, H. Xu, L. Changolkar, N. El Mammeri, V.M.-Y. Lee, and M. Hong, “Fluent Molecular Mixing of Tau Isoforms in Alzheimer’s Disease Neurofibrillary Tangles“, Nat. Commun. 13, 2967 (2022).
223
J. Medeiros-Silva, N. Somberg, H. K. Wang, M.J. McKay, V.S. Mandala, A.J. Dregni, and M. Hong, “pH- and Calcium-Dependent Aromatic Network in the SARS-CoV-2 Envelope Protein“, J. Am. Chem. Soc., 144, 6839-6850 (2022).
222
M. Sutherland, N. Tran, and M. Hong, “Clustering of the Influenza M2 Protein in Lipid Bilayers Investigated by 19F Solid-State NMR”, Biochim. Biophys. Acta, 1864, 183909 (2022).
221
H.S. Temple, P. Phyo, W. Yang, A. Echevarria-Poza, J. Lyczakowski, I. Yakunin, R. Dupree, A. Orellana, M. Hong and P. Dupree, “Discovery of putative Golgi S-Adenosyl methionine transporters reveals the importance of plant cell wall polysaccharide methylation”, Nat. Plants, 8, 656 (2022).
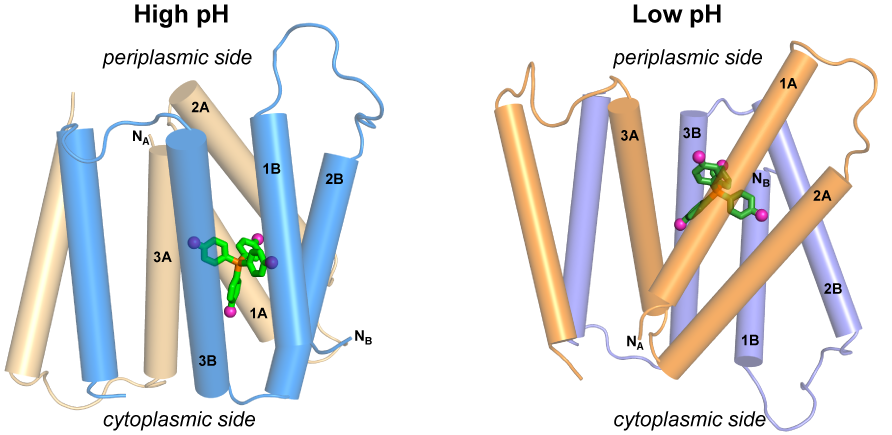
220
A.A. Shcherbakov, P.J. Spreacker, A.J. Dregni, K.A. Henzler-Wildman and M. Hong, “High-pH structure of EmrE reveals the mechanism of proton-coupled transport”, Nat. Commun., 13, 991 (2022).
219
P. Duan, K. J. Chen, G. Wijegunawardena, A.J. Dregni, H.K. Wang, H. Wu, and M. Hong, “Binding Sites of a Positron Emission Tomography Imaging Agent in Alzheimer’s b-Amyloid Fibrils Studied Using 19F Solid-State NMR”, J. Am. Chem. Soc. 144, 1416-1430 (2022).
218
A.A. Shcherbakov$, J. Medeiros-Silva$, N. Tran, M.D. Gelenter, and M. Hong, “From Angstroms to Nanometers: Measuring Interatomic Distances in Solid-State NMR”, Chemical Reviews, 122, 9848-9879 (2022).
2021
217
N. Tran, Y. Oh, M. Sutherland, Q. Cui, and M. Hong, “Cholesterol-mediated clustering of the HIV fusion Protein gp41 in lipid bilayers”, J. Mol. Biol., 434 (2), 167345 (2021).
216
M.D. Gelenter, K.J. Chen and M. Hong, “Off-Resonance 13C-2H REDOR NMR for Site-Resolved Studies of Molecular Motion”, J. Biomol. NMR, 75, 335-345 (2021).
215
M. Sutherland, B. Kwon, and M. Hong, “Interactions of HIV gp41’s membrane-proximal external region and transmembrane domain with lipid membranes from 31P NMR”, Biochim. Biophys. Acta, 1863, 183723 (2021).
214
M.D. Gelenter, A.J. Dregni, P. Duan and M. Hong, “Structurally based design of glucagon mutants that inhibit fibril formation”, Biochemistry, 60, 2033-2043 (2021).
213
P. Duan, S.J. Kaser, J. J. Lyczakowski, P. Phyo, T. Tryfona, P. Dupree, and M. Hong “Developmental-Stage Dependent Xylan Structure and Dynamics in Brachypodium Cell Walls”, ACS Omega, 6, 15460-15471 (2021).
212
N.H. Somberg, M.D. Gelenter and M. Hong, “Comparative Analysis of 13C Chemical Shifts of b-sheet Amyloid Proteins and Membrane Proteins”, J. Biomol. NMR, 75, 151-166 (2021).
211
A.J. Dregni, H. K. Wang, H. Wu, P. Duan, W.F. DeGrado and M. Hong, “Inclusion of the C-Terminal Domain in the Tau Amyloid Fibril Core Revealed by Solid-State NMR Spectroscopy”, J. Am. Chem. Soc., 143, 7839-7851 (2021).
210
M.D. Gelenter$,V.S. Mandala$, M.J.M. Niesen$, D.A. Sharon$, A. J. Dregni, A. P. Willard, and M. Hong, “Water orientation and dynamics in the closed and open influenza B virus M2 proton channels”, Commun. Biol., 4, 338 (2020).
Featured in MIT News: Chemists gain new insights into the behavior of water in an influenza virus channel: Research on how water behaves in a proton channel provides possible new avenues for flu treatment.
209
M.R. Elkins, A. Bandara, G.A. Pantelopulous, J. E. Straub, and M. Hong, Direct Observation of Cholesterol Dimers and Tetramers in Lipid Bilayers, J. Phys. Chem. B., 125 (7), 1825-1837 (2021).
208
B. Reif, S. E. Ashbrook, L. Emsley, and M. Hong, “Solid-State NMR Spectroscopy”, Nature Reviews Methods Primers, 1, 2 (2021).
207
A.A. Shcherbakov, G. S. Hisao, V.S. Mandala, N. E. Thomas, M. Soltani, E. A. Salter, J. H. Davis Jr., K. Henzler-Wildman and M. Hong, “Drug-Binding Structure and Drug Dynamics of the Bacterial Transporter EmrE in Lipid Bilayers”, Nat. Comm., 12, 172 (2021).
2020
206
V.S. Mandala, M.J. McKay, A.A. Shcherbakov, A.J. Dregni, A. Kolocouris, and M. Hong, “Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers”, Nat. Struct. Mol. Biol. 27, 1202-1208 (2020)
Featured in MIT News: Chemists discover the structure of a key coronavirus protein: The protein, which acts as an ion channel, could be a target for new drugs against the SARS-CoV-2 virus.
Featured in C&EN: Structure of SARS-CoV-2 envelope protein solved by NMR: 3-D structure of the E protein from the COVID-19-causing virus unveiled.
205
V.S. Mandala, D. Loh, S.M. Shepard, M.B. Geeson, I.V. Sergeyev, D.G. Nocera, C. C. Cummins, and M. Hong, “Bacterial phosphate granules contain cyclic polyphosphates: Evidence from 31P solid-state NMR”, J. Am. Chem. Soc. 142, 18407-18421 (2020)
204
B. Kwon, T. Mandal, M. R. Elkins, Y. Oh, Q. Cui and M. Hong, “Cholesterol Interactions with the Trimeric HIV Fusion Protein gp41 in Lipid Bilayers Investigated by Solid-State NMR Spectroscopy and Molecular Dynamics Simulations”, J. Mol. Biol., 432, 4705-4721 (2020).
203
A.J. Dregni$, P. Duan$, and M. Hong, “Hydration and Dynamics of Full-Length Tau Amyloid Fibrils Investigated by Solid-State NMR”, Biochemistry, 59, 2237-2248 (2020).
202
M.D. Gelenter, A.J. Dregni, and M. Hong, “Pulsed Third-Spin-Assisted Recoupling NMR for Obtaining Long-Range 13C-13C and 15N-13C Distance Restraints”, J. Phys. Chem., 124, 7138-7151 (2020).
201
A.A. Shcherbakov, M. Roos, B. Kwon, and M. Hong, “Two-dimensional 19F-13C correlation NMR for 19F resonance assignment of fluorinated proteins”, J. Biomol. NMR, 74, 193-204 (2020).
200
V.S. Mandala, A.R. Loftis, A.A. Shcherbakov, B.L. Pentelute, and M. Hong, “Atomic structures of closed and open influenza B M2 proton channel reveal the conduction mechanism”, Nat. Struct. Mol. Biol., 27 (2), 160-167 (2020).
Featured in MIT News: Chemists unveil the structure of an influenza B protein: Findings could help researchers design drugs to treat influenza B infections.
2019
199
M. Lee$, C. A. Morgan$, and M. Hong, “Fully hydrophobic HIV gp41 adopts a hemifusion-like conformation in phospholipid bilayers ”, J. Biol. Chem., 294, 14732-14744 (2019).
198
P. Phyo and M. Hong, “Fast MAS ¹H-¹³C correlation NMR for structural investigations of plant cell walls”, J. Biomol. NMR, 73 (12), 661-674 (2019).
Featured in MIT News: Pyae Phyo: From Myanmar to NMR — PhD candidate’s journey to the center of the plant cell wall relies on nuclear magnetic resonance technology.
197
A.J. Dregni$, V.S. Mandala$, H. Wu$, M.R. Elkins, H.K. Wang, I. Hung, W.F. DeGrado, and M. Hong, “In vitro 0N4R tau fibrils contain a monomorphic β-sheet core enclosed by dynamically heterogeneous fuzzy coat segments”, Proc. Natl. Acad. Sci. USA, 116 (33), 16357-16366 (2019).
196
M.D. Gelenter, K.J. Smith$, S.Y. Liao$, V.S. Mandala, A.J. Dregni, M.S. Lamm, Y. Tian, W. Xu, D.J. Pochan, T.J. Tucker, Y. Su, and M. Hong, “The peptide hormone glucagon forms amyloid fibrils with two coexisting β-strand conformations”, Nat. Struct. Mol. Biol., 26, 592-598 (2019).
Featured in MIT News: Chemists discover structure of glucagon fibrils: Study may be a step toward shelf-stable versions of the hormone, which is used to control diabetes.
195
B. Kwon$, M. Roos$, V.S. Mandala, A.A. Shcherbakov, and M. Hong, “Elucidating Relayed Proton Transfer Through a His-Trp-His Triad of a Transmembrane Proton Channel by Solid-State NMR”, J. Mol. Biol., 431, 2554-2566 (2019).
194
A.A. Shcherbakov, V.S. Mandala and M. Hong, “High-Sensitivity Detection of Nanometer 1H-19F Distances for Protein Structure Determination by 1H-Detected Fast MAS NMR”, J. Phys Chem. B, 123, 4387-4391 (2019).
193
M.R. Elkins and M. Hong, “Elucidating Ligand-Bound Structures of Membrane Proteins Using Solid-State NMR Spectroscopy”, Curr. Opin. Struct. Biol., 57, 103-109 (2019).
192
V.S. Mandala, S.Y. Liao, M.D. Gelenter and M. Hong, “Backbone Conformation of the Influenza Virus B M2 Protein in Lipid Bilayers from Solid-State NMR”, Sci. Rep., 9, 3725 (2019).
191
P. Phyo, Y. Gu and M. Hong, “Impact of acid pH on plant cell wall polysaccharide structure and dynamics: insights into the mechanism of acid growth of plants from solid-state NMR”, Cellulose, 26, 291-304 (2019).
190
V.S. Mandala and M. Hong, “High-sensitivity protein solid-state NMR spectroscopy”, Curr. Opin. Struc. Biol., 58, 183-190 (2019).
189
F. Li, P. Phyo, J. Jacobowitz, M. Hong, and J. K. Weng, “The molecular structure of plant sporopollenin”, Nature Plants, 5, 41-46 (2019).
Featured in MIT News: Cracking a tough case: Whitehead Institute and MIT researchers uncover the detailed molecular structure of the sporopollenin polymer, an inert material key for the emergence of land plants.
2018
188
M.R. Elkins, I.V. Sergeyev, and M. Hong, “Determining Cholesterol Binding to Membrane Proteins by Cholesterol 13C Labeling in Yeast and Dynamic Nuclear Polarization NMR”, J. Am. Chem. Soc., 140, 15437-15449 (2018).
187
M.K. Roos$, V.S. Mandala$, and M. Hong, “Determination of Long-Range Distances by Fast Magic-Angle-Spinning Radiofrequency-Driven 19F-19F Dipolar Recoupling NMR”, J. Phys. Chem. B, 122 (40), 9302-9313 (2018). ($ indicates equal contribution)
186
M.D. Gelenter and M. Hong, “Efficient 15N-13C polarization transfer by third-spin assisted pulsed cross polarization for protein structure determination by solid-state NMR”, J. Phys. Chem. B, 122 (35), 8367-8379 (2018).
185
B. Kwon$, M. Lee$, A.J. Waring, and M. Hong, “Oligomeric Structure and Three-Dimensional Fold of the HIV gp41 MPER and Transmembrane Domain in Phospholipid Bilayers”, J. Am. Chem. Soc., 140 (26), 8246-8259 (2018).
184
A. A. Shcherbakov, M. Hong. “Rapid Measurement of Long-Range Distances in Proteins by Multidimensional 13C-19F REDOR NMR Under Fast Magic-Angle Spinning“, J. Biomol. NMR 71, 31-43 (2018). doi: https://doi.org/10.1007/s10858-018-0187-0.
183
M. Roos, T. Wang, A. A. Shcherbakov, M. Hong. “Fast Magic-Angle-Spinning 19F Spin Exchange NMR for Determining Nanometer 19F-19F Distances in Proteins and Pharmaceutical Compounds“, J. Phys. Chem. B 122 (11), 2900-2911 (2018). doi: 10.1021/acs.jpcb.8b00310.
182
P. Phyo, T. Wang, H. O’Neil, and M. Hong, “Direct Determination of Hydroxymethyl Conformations of Plant Cell Wall Cellulose Using 1H Polarization Transfer Solid-State NMR”, Biomacromolecules, 19 (5), 1485-1497 (2018). doi: 10.1021/acs.biomac.8b00039
181
V. S. Mandala, J. K. Williams, and M. Hong, “Structure and Dynamics of Membrane Proteins from Solid-State NMR”, Annu. Rev. Biophys. 47, 201-222 (2018).
180
S. Y. Liao, M. W. Lee, and M. Hong, “Interplay Between Membrane Curvature and Protein Conformational Equilibrium Investigated by Solid-State NMR”, J. Struct. Biol. Epub ahead of print (2018). doi: 10.1016/j.jsb.2018.02.007.
179
V. S. Mandala, M. D. Gelenter, M. Hong, “Transport-Relevant Protein Conformational Dynamics and Water Dynamics on Multiple Time Scales in an Archetypal Proton Channel: Insights from Solid-State NMR”, J. Am. Chem. Soc. 140, 1514-1524 (2018). doi: 10.1021/jacs.7b12464.
178
M. Lee, H. W. Yao, B. Kwon, A. J. Waring, P. Ruchala, C. Singh and M. Hong, “Conformation and Trimer Association of the Transmembrane Domain of the Parainfluenza Virus Fusion Protein in Lipid Bilayers From Solid-State NMR: Insights Into the Sequence Determinants of Trimer Structure and Fusion Activity”, J. Mol. Biol. 430, 695-709 (2018).
177
H. Yang, T. Wang, D. Oehme, L. Petridis, M. Hong and J. D. Kubicki, “Structural factors affecting 13C NMR chemical shifts of cellulose – A computational study”, Cellulose, 25, 23-36 (2018).
2017
176
P. Phyo, T. Wang, S. Kiemle, Hugh O’Neill, Sai V. Pingali, M. Hong, and D.J. Cosgrove, “Gradients in wall mechanics and polysaccharides along growing Arabidopsis inflorescence stems”, Plant Physiology , 175, 1593-1607 (2017).
175
M.R. Elkins, J. K. Williams, M. D. Gelenter, P. Dai, B.S. Kwon, I.V. Sergeyev, B.L. Pentelute, and M. Hong, “Cholesterol Binding Site of the Influenza M2 Protein in Lipid Bilayers from Solid-State NMR”, Proc. Natl. Acad. Sci. USA 114 (49), 12946-12951 (2017).
Featured in The Boston Globe: MIT researchers gain new insight on how flu spreads from cell to cell in the body
Featured in MIT News: Cholesterol helps flu virus escape through host cell’s membrane: Study could shed light on how many other proteins bind with membrane cholesterol.
174
J. K. Williams, A. A. Shcherbakov, J. Wang, M. Hong, “Protonation Equilibria and Pore-opening Structure of the Dual-histidine Influenza B Virus M2 Transmembrane Proton Channel from Solid-state NMR“, J. Biol. Chem. 292 (43), 17876-17884 (2017).
173
P. Phyo, T. Wang, C. Xiao, C. Anderson, and M. Hong, “Effects of pectin mutations on the structure of plant primary cell walls: insights from solid-state NMR”, Biomacromolecules 18, 2937-2950 (2017).
172
P. Dai, J. K. Williams, C. Zhang, M. Welborn, J. J. Shepherd, T. Zhu, T. Van Voorhis, M. Hong, B. L. Pentelute, “A Structural and Mechanistic Study of π-Clamp-Mediated Cysteine Perfluoroarylation“, Sci. Rep. 7, 7954 (2017).
171
M. D. Gelenter, T. Wang, S. Liao, H. O’Neill, M. Hong, “2H-13C Correlation Solid-State NMR for Investigating Dynamics and Water Accessibilities of Proteins and Carbohydrates“, J. Biomol. NMR. 68 (4), 257-270 (2017).
170
T. Wang and M. Hong, “Structure and Dynamics of Polysaccharides in Plant Cell Walls From Solid-State NMR”, Chapter 13, NMR in Glycoscience and Glycotechnology Royal Society of Chemistry (2017).
169
V. S. Mandala, S. Liao, B. Kwon and M. Hong, “Structural Basis of Inward Rectification in the Influenza M2 Proton Channel from Solid-State NMR”, J. Mol. Biol. 429, 2192-2210 (2017).
168
M. Lee, T. Wang, O. V. Makhlynets, Y. Wu, N. Polizzi, H. Wu, J. Stöhr, I. V. Korendovych, W. F. DeGrado, and M. Hong, “Zinc-Binding Structure of a Catalytic Amyloid from Solid-State NMR Spectroscopy”, Proc. Natl. Acad. Sci. USA. 114 (24), 6191-6196 (2017).
Featured in MIT News: Chemists reveal amyloid structure: Discovery of how amyloids bind metal ions sheds light on protein function.
167
T. Wang, H. Jo, W.F. DeGrado, M. Hong, “Water Distribution, Dynamics and Interactions with Alzheimer’s Beta-Amyloid Fibrils Investigated by Solid-State NMR“, J. Am. Chem. Soc. 139 (17), 6242-6252 (2017).
166
D. Chen, M.D. Gelenter, M. Hong, R.E. Cohen, G.H. McKinley, “Icephobic Surfaces Induced by Interfacial Non-frozen Water“, ACS Appl. Mater. Interfaces 9 (4), 4202-4214 (2017).
2016
165
R. Liang, J.M. Swanson, J.J. Madsen, M. Hong, W.F DeGrado and G.A. Voth, “Acid Activation Mechanism of the Influenza A M2 Proton Channel“, Proc. Natl. Acad. Sci. USA. 113, E6955-E6964 (2016).
164
H. Yao, M. Lee, S. Liao and M. Hong, “Solid-State NMR Investigation of the Structural Topology and Lipid Interactions of a Viral Fusion Protein Chimera Containing the Fusion Peptide and Transmembrane Domain“, Biochemistry 55, 6787-6800 (2016).
163
T. Wang, Y. Chen, A. Tabuchi, D.J. Cosgrove and M. Hong, “The Target of Beta-Expansion EXPB1 in Maize Cell Walls from Binding and Solid-State NMR Studies”, Plant Physiol. 172, 2107-2119 (2016).
162
B. Kwon and M. Hong, “The Influenza M2 Ectodomain Regulates the Conformational Equilibria of the Transmembrane Proton Channel: Insights from Solid-State NMR“, Biochemistry 55, 5387-5397 (2016).
161
T. Wang, P. Phyo, M. Hong, “Multidimensional Solid-State NMR Spectroscopy of Plant Cell Walls“, Solid State Nucl. Magn. Reson. 78, 56-63 (2016).
160
T. Wang, H. Yang, J. D. Kubicki and M. Hong, “Cellulose Structural Polymorphism in Plant Primary Cell Walls Investigated by High-Field 2D Solid-State NMR Spectroscopy and Density Functional Theory Calculations“,Biomacromolecules, 17, 2210-2222 (2016).
159
M.R. Elkins, T. Wang, M. Nick, H. Jo, T. Lemmin, S. Prusiner, W.F. DeGrado, J. Stohr, M. Hong, “Structural Polymorphism of Alzheimer’s Beta-Amyloid Fibrils as Controlled by an E22 Switch: A Solid-State NMR Study“, J. Am. Chem. Soc. 138, 9840-9852 (2016).
158
J.K. Williams, D. Tietze, M. Lee, J. Wang, M. Hong, “Solid-State NMR Investigation of the Conformation, Proton Conduction, and Hydration of the Influenza B Virus M2 Transmembrane Proton Channel“, J. Am. Chem. Soc. 138, 8143-8155 (2016).
157
S. Liao, M. Lee, T. Wang. I.V. Sergeyev and M. Hong, “Efficient DNP NMR of Membrane Proteins: Sample Preparation Protocols, Sensitivity, and Radical Location“, J. Biomol. NMR 64, 223-237 (2016).
156
K.J. Fritzsching, M. Hong and K. Schmidt-Rohr, “Conformationally selective multidimensional chemical shift ranges in proteins from a PACSY database purged using intrinsic quality criteria“, J. Biomol. NMR 64, 115-130 (2016).
155
T. Wang and M. Hong, “Solid-State NMR Investigations of Cellulose Structure and Interactions with Matrix Polysaccharides in Plant Primary Cell Walls“, J. Exp. Bot. 67, 503-514 (2016).
2015
154
J.K. Williams, K. Schmidt-Rohr and M. Hong, “Aromatic Spectral Editing Techniques for Magic-Angle-Spinning Solid-State NMR Spectroscopy of Uniformly 13C-Labeled Proteins“, Solid State Nucl. Magn. Reson. 72, 118-126 (2015).
153
H. Yao, M.W. Lee, A.J. Waring, G.C.L. Wong, and M. Hong, “A Viral Fusion Protein Transmembrane Domain Adopts β-Strand Structure to Facilitate Membrane Topological Changes for Virus-Cell Fusion“, Proc. Natl. Acad. Sci. USA, 112, 10926-10931 (2015).
152
P.B. White and M. Hong, “15N and 1H Solid-State NMR Investigation of a Canonical Low-Barrier Hydrogen-Bond Compound: 1,8-bis(dimethylamino) Naphthalene”, J. Phys. Chem. B, 119, 11581-11589 (2015).
151
T. Wang, Y.B. Park, D.J. Cosgrove, and M. Hong, “Cellulose-Pectin Spatial Contacts are Inherent to Never-Dried Arabidopsis thaliana Primary Cell Walls: Evidence from Solid-State NMR”, Plant Physiol. 168, 871-884 (2015).
150
B.S. Kwon, D. Tietze, P.B. White, S. Liao and M. Hong, “Chemical Ligation of the Influenza M2 Protein for Solid-State NMR Characterization of the Cytoplasmic Domain”, Protein Sci. 24, 1087-1099 (2015).
149
S. Liao, Y. Yang, D. Tietze and M. Hong, “The Influenza M2 Cytoplasmic Tail Changes the Proton-Exchange Equilibria and the Backbone Conformation of the Transmembrane Histidine Residue to Facilitate Proton Conduction”, J. Am. Chem. Soc. 137, 18, 6067-6077 (2015).
148
Y. Yang, H. Yao and M. Hong, “Distinguishing Bicontinuous Lipid Cubic Phases from Isotropic Membrane Morphologies Using 31P Solid-State NMR Spectroscopy“, J. Phys. Chem. B 119, 4993-5001 (2015).
147
T. Wang and M. Hong, “Investigation of the Curvature Induction and Membrane Localization of the Influenza Virus M2 Protein Using Static and Off-Magic-Angle-Spinning Solid-State Nuclear Magnetic Resonance of Oriented Bicelles“, Biochemistry, 54, 2214-2226 (2015).
146
T. Wang, J.K. Williams, K. Schmidt-Rohr and M. Hong, “Relaxation-Compensated Difference Spin Diffusion NMR for Detecting 13C-13C Long-Range Correlations in Proteins and Polysaccharides“, J. Biomol. NMR 61, 97-107 (2015).
2014
145
145. N. Joh, T. Wang, M. Bhate, R. Acharya, Y. Wu, M. Grabe*, M. Hong*, G. Grigoryan* and W.F. DeGrado*, “De Novo Design of a Transmembrane Zn(II) Transporting Four-Helix Bundle”, Science 346, 1520-1524 (2014). News and Events, Chemistry Department C&EN Science and Technology News, Dec 22, 2014.
144
J.K. Williams and M. Hong, “Probing Membrane Protein Structure Using Water Polarization Transfer Solid-State NMR”, J. Magn. Reson. 247, 118-127 (2014).
143
M. Lee and M. Hong, “Cryoprotection of Lipid Membranes for High-Resolution Solid-State NMR Studies of Membrane Peptides and Proteins at Low Temperature”, J. Biomol. NMR 59, 263-277 (2014).
142
P.B. White, T. Wang, Y.B. Park, D.C. Cosgrove and M. Hong, “Hydration of Cellulose and Matrix Polysaccharides in the Primary Cell Wall of Arabidopsis Thaliana from Spin Diffusion NMR”, J. Am. Chem. Soc.136, 10399-409 (2014).
141
T. Wang, O. Zabotina, and M. Hong, “Structure and Dynamics of Brachypodium Primary Cell Wall Polysaccharides from Solid-State NMR Spectroscopy”, Biochemistry 53, 2840-2854 (2014).
140
H. Yao and M. Hong, “Conformation and Lipid Interaction of the Fusion Peptide of the Paramyxovirus PIV5 in Anionic and Negative-Curvature Membranes From Solid-State NMR”, J. Am. Chem. Soc. 136, 2611-2624 (2014).
2013
139
Y. Yang, K.J. Fritzsching, and M. Hong, “Resonance Assignment of Disordered Proteins Using a Multi-Objective Non-Dominated Sorting Genetic Algorithm”, J. Biomol. NMR 57, 281-96 (2013).
138
S. Liao, K.J. Fritzsching, and M. Hong, “Conformational analysis of the full-length M2 protein of the influenza A virus using solid-state NMR”, Protein Sci. 22, 1623-38 (2013).
137
T. Wang, Y.B. Park, M.A. Caporini, M. Rosay, D.J. Cosgrove and M. Hong, “Sensitivity-Enhanced Solid-State NMR Detection of Expansin’s target in Plant Cell Walls”, Proc. Natl. Acad. Sci. U.S.A. 110, 16444-9 (2013).
136
B. Kwon, A.J. Waring and M. Hong, “A 2H Solid-State NMR Study of Lipid Clustering by Cationic Antimicrobial and Cell-Penetrating Peptides in Model Bacterial Membranes”, Biophys. J. 105, 2333-42 (2013).
135
R.L. Johnson, J.M. Anderson, B.H. Shanks, X. Fang, M. Hong, K. Schmidt-Rohr, “Spectrally edited 2D 13C-13C NMR spectra without diagonal ridge for characterizing 13C-enriched low-temperature carbon materials”, J. Magn. Reson. 234C, 112-124 (2013).
134
J.K. Williams, D. Tietze, J. Wang, Y. Wu, W.F. DeGrado and M. Hong, “Drug-Induced Conformational and Dynamical Changes of the S31N Mutant of the Influenza M2 Proton Channel Investigated by Solid-State NMR”, J. Am. Chem. Soc. 135, 9885-9897 (2013).
133
K.J. Fritzsching, Y. Yang, K. Schmidt-Rohr, and M. Hong, “Practical Use of Chemical Shift Databases for Protein Solid-State NMR: 2D Chemical Shift Maps and Amino-Acid Assignment with Secondary-Structure Information”, J. Biomol. NMR 56, 155-167 (2013).
132
T. Wang, H. Yao and M. Hong, “Determining the Depth of Insertion of Dynamically Invisible Membrane Peptides by Gel-Phase 1H Spin Diffusion Heteronuclear Correlation NMR”, J. Biomol. NMR 56, 139-148 (2013).
131
J. Williams, Y. Zhang, K. Schmidt-Rohr, and M. Hong, “Solid-state NMR investigation of the pH-dependent structure and dynamics of the gating residue of the influenza M2 proton channel”, Biophys. J. 104, 1698-1708 (2013). New and Notable commentary on 1639-1640.
130
M. Hong and K. Schmidt-Rohr, “Magic-Angle-Spinning NMR Techniques for Measuring Long-Range Distances in Biological Macromolecules”, Acct. Chem. Res. 46, 2154-63 (2013).
129
Su Y, Li S, and Hong M. “Cationic membrane peptides: atomic-level insight of structure-activity relationships from solid-state NMR”, Amino Acids 44, 821-833 (2013).
128
H. Yao, and M. Hong, “Membrane-Dependent Conformation, Dynamics, and Lipid Interactions of the Fusion Peptide of the Paramyxovirus PIV5 From Solid-State NMR”, J. Mol. Biol. 425, 563-576 (2013).
2012
127
T. Wang, O. Zabotina, and M. Hong, “Pectin-cellulose and protein-polysaccharide interactions in Arabidopsisprimary cell walls by 2D 13C Correlation NMR”, Biochemistry 51, 9846-9856 (2012).
126
T. Wang, L. Widanapathirana, Y. Zhao and M. Hong, “Aggregation and Dynamics of Oligocholate Transporters in Phospholipid Bilayers Revealed by Solid-State NMR Spectroscopy”, Langmuir 28, 17071-17078 (2012).
125
K. Schmidt-Rohr, K. J. Fritzsching, S. Liao, M. Hong, “Spectral Editing of Two-Dimensional Magic-Angle-Spinning Solid-State NMR Spectra for Protein Resonance Assignment and Structure Determination”, J. Biomol. NMR 54, 343-353 (2012).
124
M. Hong and W.F. DeGrado, “Structural Basis For Proton Conduction and Inhibition by the Influenza M2 Protein”, Invited review, Protein Sci. 21, 1620-1633 (2012).
123
S. Li, Y. Su, and M. Hong, “Intramolecular 1H-13C distance measurement in uniformly 13C, 15N labeled peptides by solid-state NMR”, Solid State Nucl. Magn. Reson. 45-46, 51-58 (2012).
122
M. Hong, K.J. Fritzsching, and J. K. Williams, “Hydrogen-Bonding Partner of the Proton-Conducting Histidine in the Influenza M2 Proton Channel Revealed From 1H Chemical Shifts”, J. Am. Chem. Soc. 134, 14753-5 (2012).
121
Y. Su, F. Hu and M. Hong, “Paramagnetic Cu(II) For Probing Membrane Protein Structure and Function: Inhibition Mechanism of the Influenza M2 Proton Channel”, J. Am. Chem. Soc. 134, 8693-8702, 2012.
120
M. Dick-Perez, T. Wang, A. Salazar, O. Zabotina, and M. Hong, “Multidimensional Solid-State NMR Studies of the Structure and Dynamics of Pectic Polysaccharides in Uniformly 13C-Labeled Arabidopsis Primary Cell Walls”,Magn. Reson. Chem. 50, 539-550 (2012).
119
D.M. Harris, K. Corbin, T. Wang, R. Gutierrez, A.L. Bertolo, C. Petti, D.M. Smilgies, J. M. Estevez, D. Bonetta, B. Urbanowicz, D. Ehrhardt, C.R. Somerville, J. K.C. Rose, M. Hong, S. DeBolt, “Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase”, Proc. Natl. Acad. Sci. U.S.A. 109, 4098-4103 (2012).
118
T. Wang, S.D. Cady, and M. Hong, “NMR Determination of Protein Partitioning into Membrane Domains with Different Curvatures and Application to the Influenza M2 Protein”, Biophys. J. 102, 787-794 (2012).
117
F. Hu, K. Schmidt-Rohr and M. Hong, “NMR Detection of pH-Dependent Histidine-Water Proton Exchange Reveals the Conduction Mechanism of a Transmembrane Proton Channel”, J. Am. Chem. Soc. 134, 3703-3713 (2012). Cover of JACS volume 134, issue 8.
116
M. Hong, Y. Zhang and F. Hu, “Membrane protein structure and dynamics from NMR spectroscopy”, Review,Annu. Rev. Phys. Chem. 63, 1-24. (2012).
2011
115
Y. Su and M. Hong, “Conformational disorder of membrane peptides investigated from solid-state NMR line widths and line shapes”, J. Phys. Chem. 115, 10758-10767 (2011).
114
J. Wang, C. Ma, G. Fiorin, V. Carnevale, T. Wang, F. Hu, R.A. Lamb, M.L. Klein, L.H. Pinto, M. Hong, and W.F. DeGrado, “Molecular Dynamics Simulation Directed Rational Design of Inhibitors Targeting Drug-Resistant Mutants of Influenza A Virus M2”, J. Am. Chem. Soc. 133, 12834-12841 (2011).
113
S.D. Cady, T. Wang, and M. Hong, “Membrane-Dependent Effects of a Cytoplasmic Helix on the Structure and Drug Binding of the Influenza M2 Protein”, J. Am. Chem. Soc. 133, 11572-11579 (2011).
112
M. Hong and Y. Su, “Structure and Dynamics of Cationic Membrane Peptides and Proteins: Insights from Solid-State NMR”, Protein Sci. review, 20, 641-655 (2011).
111
Y. Su, A.J. Waring, P. Ruchala, and M. Hong, “Structure of beta-hairpin Antimicrobial Peptides in Lipopolysaccharide Membranes: Mechanism of Gram Selectivity Obtained from Solid-State Nuclear Magnetic Resonance”, Biochemistry 50, 2072-2083 (2011).
110
S.D. Cady, J. Wang, Yibing Wu, W.F. DeGrado, and M. Hong, “Specific binding of adamantane drugs and direction of their polar amines in the pore of the influenza M2 transmembrane domain in lipid bilayers and dodecylphosphocholine micelles determined by NMR spectroscopy”, J. Am. Chem. Soc. 133, 4274-4284 (2011).
109
M. Dick-Perez, Y. Zhang, J. Hayes, A. Salazar, O.A. Zabotina and M. Hong, “Structure and Interactions of Plant Cell-Wall Polysaccharides by 2D and 3D Magic-Angle-Spinning Solid-State NMR”, Biochemistry 50, 989-1000 (2011).
108
S. Li and M. Hong, “Protonation, Tautomerization and Rotameric Structure of Histidine: A Comprehensive Study through High Resolution Solid-state NMR”, J. Am. Chem. Soc. 133, 1534-1544 (2011).
107
F. Hu, W. Luo, S.D. Cady and M. Hong, “Conformational Plasticity of the Influenza A M2 Transmembrane Peptide in Lipid Bilayers Under Varying pH, Drug Binding and Membrane Thickness”, Biochim. Biophys. Acta1808, 415-423 (2011).
2010
106
Y. Zhang and M. Hong, “Membrane-bound structure and topology of a human alpha-defensin indicates dimer pore as the mechanism of antimicrobial activity”, Biochemistry 49, 9770-9782 (2010).
105
F. Hu, W. Luo, and M. Hong, “Mechanisms of proton conduction and gating by influenza M2 proton channels from solid-state NMR”, Science 330, 505-508 (2010).
104
Y. Su, A.J. Waring, P. Ruchala, and M. Hong, “Membrane-bound dynamic structure of an Arginine-rich cell-penetrating peptide, the protein transduction domain of HIV TAT, from solid-state NMR”, Biochemistry 49, 6009-6020 (2010).
103
Y. Su, W.F. DeGrado and M. Hong, “Orientation, dynamics and lipid interaction of an antimicrobial arylamide in lipid bilayers investigated by 19F and 31P solid-state NMR”, J. Am. Chem. Soc. 132, 9197-9205 (2010).
102
T. Doherty*, Y. Su* and M. Hong, “High-Resolution Orientation and Depth of Insertion of the Voltage-Sensing S4 Helix of a Potassium Channel in Lipid Bilayers”, J. Mol. Biol. 401, 642-652 (2010). (* indicates equal contribution)
101
S. Li, Y. Su, W. Luo, M. Hong, “Water-protein interactions of an arginine-rich membrane peptide in lipid bilayers investigated by solid-state nuclear magnetic resonance spectroscopy”. J. Phys. Chem. B 114, 4063-4069.
100
W. Luo and M. Hong, “Conformational Changes of an Ion Channel Detected Through Water-Protein Interactions Using Solid-State NMR”, J. Am. Chem. Soc. 132, 2378-2384 (2010).
99
S.D. Cady, K. Schmidt-Rohr, J. Wang, C.S. Soto, W.F. DeGrado, and M. Hong, “Structure of the amantadine binding site of influenza M2 proton channels In lipid bilayers”, Nature 463, 689-692 (2010).
98
S. Li, Y. Zhang, and M. Hong, “3D 13C-13C-13C Correlation NMR for De Novo Distance Determination of Solid Proteins and Application to a Human Alpha Defensin”, J. Magn. Reson. 202, 203-210 (2010).
97
Y. Zhang, T. Doherty, J. Li, W. Lu, C. Barinka, J. Lubkowski and M. Hong, “Resonance Assignment and Three-Dimensional Structure Determination of a Human Alpha-Defensin, HNP-1, by Solid-State NMR”, J. Mol. Biol. 397, 408-422 (2010).
2009
96
S.D. Cady, W. Luo, F. Hu, and M. Hong, “Structure and function of the influenza M2 proton channel”, Current Topic, Biochemistry 48, 7356-7364 (2009).
95
S.D. Cady and M. Hong, “Effects of Amantadine Binding on the Dynamics of Bilayer-Bound Influenza A M2 Transmembrane Peptide Studied by NMR Relaxation”, J. Biomol. NMR 45, 185-196. (2009).
94
T. Doherty and M. Hong, “High-Resolution Solid-State NMR of Anisotropically Mobile Molecules Under Very Low Power 1H Decoupling and Moderate Magic-Angle Spinning”, J. Magn. Reson. 199, 225-232 (2009).
93
M. Hong, T.V. Mishanina and S.D. Cady, “Accurate Measurement of Methyl 13C Chemical Shifts by Solid-State NMR for the Determination of Protein Sidechain Conformation: the Influenza A M2 Transmembrane Peptide as an Example”, J. Am. Chem. Soc. 131, 7806-7816 (2009).
92
Y. Su and M. Hong, “Roles of arginine and lysine residues in the translocation of a cell-penetrating peptide from13C, 31P, and 19F solid-state NMR”, Biochemistry 48, 4587-4595 (2009).
91
J. Wang, S.D. Cady, V. Balannik, L.H. Pinto, W.F. DeGrado, and M. Hong, “Discovery of Spiro-piperidine inhibitors and their modulation of the dynamics of the M2 proton channel from influenza A Virus”, J. Am. Chem. Soc. 131, 8066-8076 (2009).
90
W. Luo, S.D. Cady, and M. Hong, “Immobilization of the Influenza A M2 Transmembrane Peptide in Viral-Envelope Mimetic Lipid Membranes Detected by Solid-State NMR Spectroscopy”, Biochemistry 48, 6361-6368 (2009).
89
M. Tang. and M. Hong. “Structure and Mechanism of β-Hairpin Antimicrobial Peptides in Lipid Bilayers from Solid-State NMR Spectroscopy”, Mol. Biosys. 5, 317-322 (2009).
88
S.D. Cady, T.V. Mishanina and M. Hong, “Structure of Amantadine-Bound M2 Transmembrane Peptide of Influenza A in Lipid Bilayers from Magic-Angle-Spinning Solid-State NMR: the Role of Ser31 in Amantadine Binding,J. Mol. Biol. 385, 1127-1141 (2009).
87
M. Tang, A.J. Waring and M. Hong, “Effects of Arginine Density on the Membrane-Bound Structure of a Cationic Antimicrobial Peptide from Solid-State NMR”, Biochim. Biophys. Acta 1788, 514-521 (2009).
86
T. Doherty and M. Hong, “2D 1H-31P Solid-State NMR Studies of the Dependence of Inter-bilayer Water Dynamics on Lipid Headgroup Structure and Membrane Peptides”, J. Magn. Reson. 196, 39-47 (2009).
2008
85
Y. Su, R. Mani, T. Doherty, A.J. Waring and M. Hong, “Reversible Sheet – Turn Conformational Change of a Cell-Penetrating Peptide in Lipid Bilayers Studied by Solid-State NMR“, J. Mol. Biol. 381,1133 -1144 (2008).
84
Y. Su, R. Mani, and M. Hong, “Asymmetric Insertion of Membrane Proteins in Lipid Bilayers by Solid-State NMR Paramagnetic Relaxation Enhancement: a Cell-Penetrating Peptide Example“, J. Am. Chem. Soc. 130, 8856-8864 (2008).
83
M. Tang, A.J. Waring, and M. Hong, “Arginine Dynamics in a Membrane-Bound Cationic Beta-Hairpin Peptide from Solid-State NMR“, ChemBioChem 9, 1487-1492 (2008).
82
M. Tang, A.J. Waring, R.I. Lehrer and M. Hong, “Effects of Guanidinium-Phosphate Hydrogen Bonding on the Membrane-Bound Structure and Activity of an Arginine-Rich Membrane Peptide from Solid State NMR“, Angew. Chem. Int. Ed. Engl. 47, 3202-3205 (2008).
81
S.D. Cady and M. Hong, “Simultaneous extraction of multiple orientational constraints of membrane proteins by13C-detected N-H dipolar couplings under magic angle spinning“, J. Magn. Reson. 191, 219-225 (2008).
80
S.D. Cady and M. Hong, “Amantadine-Induced Conformational and Dynamical Changes of the Influenza M2 Transmembrane Proton Channel“, Proc. Natl. Acad. Sci. U.S.A. 105, 1483-1488 (2008).
79
T. Doherty, A.J. Waring and M. Hong, “Dynamic Structure of Disulfide-Removed Linear Analogs of Tachyplesin-I in the Lipid Bilayer from Solid-State NMR“, Biochemistry 47, 1105-1116 (2008).
2007
78
R. Mani, A.J. Waring, and M. Hong, “Conformation, Dynamics and Insertion of a Non-Cysteine-Containing Protegrin-1 Analog in Lipid Membranes from Solid-State NMR‘, ChemBioChem 8, 1877-1884 (2007).
77
M. Hong, “Structure, Topology, and Dynamics of Membrane Peptides and Proteins from Solid-State NMR Spectroscopy“, J. Phys. Chem. B feature article, 111, 10340-10351 (2007).
76
W. Luo, R. Mani and M. Hong, “Sidechain Conformation and Gating of the M2 Transmembrane Peptide Proton Channel of Influenza A Virus from Solid-State NMR“, J. Phys. Chem. B 111, 10825-10832 (2007).
75
S.D. Cady, C. Goodman, C. Tatko, W.F. DeGrado, and M. Hong, “Determining the Orientation of Uniaxially Rotating Membrane Proteins Using Unoriented Samples: a 2H, 13C, and 15N Solid-State NMR Investigation of the Dynamics and Orientation of a Transmembrane Helical Bundle“, J. Am. Chem. Soc. 129, 5719-5729 (2007).
74
M. Tang, A.J. Waring and M. Hong, “Trehalose-Protected Lipid Bilayers for Determining Membrane Protein Insertion and Topology“, J. Magn. Reson. 184, 222-227 (2007).
73
M. Tang, A.J. Waring and M. Hong, “Phosphate-Mediated Arginine Insertion Into Lipid Membranes and Pore Formation by a Cationic Membrane Peptide from Solid-State NMR“, J. Am. Chem. Soc. 129, 11438-11446 (2007).
2006
72
M. Hong and T. Doherty, “Orientation Determination of Membrane-Disruptive Proteins Using Powder Samples and Rotational Diffusion: A Simple Solid-State NMR Approach“, Chem. Phys. Lett. 432, 296-300 (2006).
71
M. Hong, “Oligomeric structure, dynamics, and orientation, of membrane proteins from solid-state NMR“,Structure, review, 14, 1731-1740 (2006).
70
T. Doherty, A.J. Waring and M. Hong, “Membrane-bound conformation and topology of Tachyplesin-1 by solid-state NMR spectroscopy“, Biochemistry, 45, 13323-13330 (2006).
69
R. Mani, S.D. Cady, M. Tang, A.J. Waring, R.I. Lehrer, and M. Hong, “Membrane-dependent oligomeric structure and pore formation of a β-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR“, Proc. Natl. Acad. Sci. U.S.A. 103, 16242-16247 (2006).
68
R. Mani, M. Tang, X. Wu, J.J. Buffy, A.J. Waring, M.A. Sherman, and M. Hong, “Membrane-Bound Dimer Structure of a β-Hairpin Antimicrobial Peptide from Rotational-Echo Double-Resonance Solid-State NMR“,Biochemistry 45, 8341-8349 (2006).
67
W. Luo and M. Hong, “Determination of the Oligomeric Number and Intermolecular Distances of Membrane Protein Assemblies by Anisotropic 1H-Driven Spin Diffusion NMR“, J. Am. Chem. Soc. 128, 7242-7251 (2006).
66
T. Doherty, A. Waring, and M. Hong, “Peptide-Lipid Interactions of the β-Hairpin Antimicrobial Peptide Tachyplesin and its Linear Derivatives from Solid-State NMR“, Biochim. Biophys. Acta 1758, 1285-1291 (2006).
65
M. Hong, “Solid-State NMR Studies of the Structure and Dynamics of Disordered and Membrane-Bound Peptides and Proteins“, Acct. Chem. Res. 39, 176-183 (2006).
64
X.L. Yao and M. Hong, “Effects of Anionic Lipid and Ion Concentrations on the Topology and Segmental Mobility of Colicin Ia Channel Domain From Solid-State NMR“, Biochemistry 45, 289-295 (2006).
63
W. Luo and M. Hong, “1D Sensitivity-Enhanced 1H Spin Diffusion Experiment for Determining Membrane Protein Topology“, Solid State Nuc. Magn. Reson. 29, 163-169 (2006).
62
M. Tang, A.J. Waring, R.I. Lehrer, and M. Hong, “Orientation of a β-hairpin Antimicrobial Peptide in Lipid Bilayers from 2D Dipolar Chemical-Shift Correlation NMR“, Biophys. J. 90, 3616-3624 (2006).
2005
61
P.A.B. Marasinghe, J.J. Buffy, K. Schmidt-Rohr and M. Hong, “Membrane Curvature Change Induced by an Antimicrobial Peptide Detected by 31P Exchange NMR“, J. Phys. Chem. B 109, 22036-44 (2005).
60
M. Tang, A.J. Waring, and M. Hong, “Intermolecular Packing and Alignment of a β-Hairpin Peptide from 2D Solid-State NMR“, J. Am. Chem. Soc. 127, 13919-13927 (2005).
59
R. Mani, A.J. Waring, R.I. Lehrer, and M. Hong, “Membrane-Disruptive Abilities of β-Hairpin Antimicrobial Peptides Correlate with Conformation and Activity: A 31P and 1H NMR Study“, Biochim. Biophys. Acta, 1716, 11-18 (2005).
58
S. Wi, H. Sun, E. Oldfield and M. Hong, “Solid-State NMR and Quantum Chemical Investigations of 13Cα Shielding Tensor Magnitudes and Orientations in Peptides: Determining φ and ψ Torsion Angles“, J. Am. Chem. Soc. 127, 6451-8 (2005).
57
W. Luo, X.L. Yao and M. Hong, “Evidence of Large Structure Rearrangement of Colicin Ia Channel Domain Due to Membrane Binding from 2D 13C Spin Diffusion NMR“, J. Am. Chem. Soc. 127, 6402-8 (2005).
56
J.J. Buffy, A.J. Waring, and M. Hong, “Determination of Peptide Oligomerization in Lipid Bilayers Using 19F Spin Diffusion NMR“, J. Am. Chem. Soc. 127, 4477-4483 (2005).
55
M. Hong and S. Wi, “Torsion Angle Determination in Biological Solids by Solid-State NMR”, chapter 4, NMR spectroscopy of Biological Solids, CRC Press, Boca Raton, 2005.
2004
54
R. Mani, J.J. Buffy, A.J. Waring, R.I. Lehrer, and M. Hong, “Solid-State NMR Investigation of the Selective Disruption of Lipid Membranes by Protegrin-1“, Biochemistry 43, 13839-48 (2004).
53
S. Wi, N. Sinha and M. Hong, “Long-range distances by heteronuclear detected 19F-1H rotational-echo double-resonance NMR“, J. Am. Chem. Soc. 126, 12754-5 (2004).
52
N. Sinha, K. Schmidt-Rohr and M. Hong, “Compensation for Pulse Imperfections in Rotational-Echo Double-Resonance NMR by Composite Pulses and EXORCYCLE“, J. Magn. Reson. 168, 358-65 (2004).
51
J.J. Buffy, M.J. McCormick, S. Wi, A. Waring, R.I. Lehrer, and M. Hong, “Selective Perturbation of Lipid Membranes by the Cyclic Antimicrobial Peptide RTD-1 Investigated by Solid-State NMR“, Biochemistry 43, 9800-9812 (2004).
50
A.T. Petkova, M. Baldus, M. Belenky, M. Hong, R.G. Griffin, J. Herzfeld, “Backbone and side chain assignment strategies for multiply labeled membrane peptides and proteins in the solid state“, J. Magn. Reson. 160: 1-12 (2003).
49
G.J. Gallagher, M. Hong, L.K. Thompson, “Solid-state NMR spin diffusion for measurement of membrane-bound peptide structure: gramicidin A“, Biochemistry 43, 7899-7906 (2004).
48
X.L. Yao and M. Hong, “Structural Distribution in an Elastin-Mimetic Peptide by Magic-Angle Spinning Solid-State NMR Spectroscopy“, J. Am. Chem. Soc. 126, 4199-4210 (2004).
47
X.L. Yao, R.A. McMillian, V.P. Conticello, and M. Hong, “Investigation of the Dynamics of an Elastin-Mimetic Polypeptide Using Solid-State NMR“, Magn. Reson. Chem. 42, 267-275 (2004).
2003
46
N. Sinha and M. Hong, “X-1H Rotational-Echo Double-Resonance NMR For Torsion Angle Determination in Peptides”, Chem. Phys. Lett. 380, 742-748 (2003).
45
J. Buffy, A.J. Waring, R. I. Lehrer, and M. Hong, “Immobilization and Aggregation of Antimicrobial Peptide Protegrin in Lipid Bilayers by Solid-State NMR”, Biochemistry, 42, 13725-34 (2003).
44
K. Schmidt-Rohr and M. Hong, “Measurements of Carbon to Amide-Proton Distances by C-H Dipolar Recoupling with 15N NMR Detection”, J. Am. Chem. Soc. 125, 5648-5649 (2003).
43
J. Buffy, T. Hong, S. Yamaguchi, A. Waring, R.I. Lehrer, and M. Hong, “Solid-State NMR Investigation of the Depth of Insertion of PG-1 in DLPC Using Paramagnetic Mn2+”, Biophys. J. 85, 2363-2373 (2003).
42
M. Hong and X.L. Yao, “Homonuclear decoupled 13C chemical shift anisotropy in multiply labeled peptides by selective-pulse solid-state NMR”, J. Magn. Reson. 160, 114-119 (2003).
41
M. Hong, D. Isailovic, R.A. McMillan, and V.P. Conticello, “Solid-state NMR chemical shift constraints for the structure of an elastin-mimetic protein”, Biopolymers, 70, 158-168, (2003).
2002
40
X.L. Yao, S. Yamaguchi and M. Hong, “Ca Chemical Shift Tensors in Helical Peptides by Dipolar-Modulated Chemical Shift Recoupling NMR”, J. Biomol. NMR 24, 51-62 (2002).
39
S. Yamaguchi, A. Waring, T. Hong, R.I. Lehrer and M. Hong, “Solid-state NMR Investigations of Peptide-Lipid Interaction and Orientation of a β-sheet antimicrobial peptide, Protegrin”, Biochemistry 41, 9852-9862 (2002).
38
M. Hong, X.L. Yao, K. Jakes and D. Huster, “Investigation of Molecular Motions by Magic-Angle Cross-Polarization NMR Spectroscopy”, J. Phys. Chem. 106, 7355-7364 (2002).
37
X.L. Yao and M. Hong, “Determination of Ca Chemical Shift Tensor Orientation in Peptides By Dipolar-Modulated Chemical Shift Recoupling Solid-State NMR”, J. Am. Chem. Soc. 124, 2730-2738 (2002).
36
D. Huster, X.L. Yao and M. Hong, “Membrane Protein Topology Probed by 1H Spin Diffusion from Lipids Using Solid-State NMR Spectroscopy”, J. Am. Chem. Soc. 124, 874-883 (2002).
35
M. Hong*, R.A. McMillan, and V.P. Conticello, “Measurement of Conformational Constraints in an Elastin-Mimetic Protein by Residue-Pair Selected Solid-State NMR”, J. Biomol. NMR 22, 175-179 (2002).
34
S. Yamaguchi and M. Hong, “Determination of Membrane Peptide Orientation by 1H-Detected 2H NMR”, J. Magn. Reson. 155, 244-250 (2002).
33
D. Huster, X.L. Yao, K. Jakes, and M. Hong, “Conformational changes of colicin Ia channel-forming domain upon membrane binding: a solid-state NMR study”, Biochim. Biophys. Acta 1561, 159-170 (2002).
2001
32
S.B. Kennedy, E. de Azevedo, W.A. Petka, D.A. Tirrell, T.P. Russell, and M. Hong, “Dynamic Structure of a Protein Hydrogel: A Solid-State NMR Study”, Macromolecules 34, 8675-8685 (2001).
31
S. Yamaguchi, D. Huster, A. Waring, R.I. Lehrer, W. Kearney, B.F. Tack, and M. Hong, “Orientation and Dynamics of an Antimicrobial Peptide in the Lipid Bilayer by Solid-State NMR”, Biophys. J. 81, 2203-2214 (2001).
30
X.L. Yao and M. Hong, “Dipolar Filtered 1H-13C Heteronuclear Correlation Spectroscopy For Resonance Assignment of Proteins”, J. Biomol. NMR 20, 263-274 (2001).
29
X. L. Yao, K. Schmidt-Rohr, and M. Hong, “Medium- and Long-Distance 1H-13C Heteronuclear Correlation NMR in Solids”, J. Magn. Reson. 149, 139-143 (2001).
28
K. Schmidt-Rohr, K. Saalwächter, S. Liu, and M. Hong, “High-Sensitivity 2H-NMR in Solids by 1H Detection”, J. Am. Chem. Soc. 123, 7168-7169 (2001).
27
D. Huster, L.S. Xiao, and M. Hong, “Solid-State NMR Investigation of the Dynamics of Colicin Ia Channel-Forming Domain”, Biochemistry 40, 7662-7674 (2001).
26
M. Hong and S. Yamaguchi, “Sensitivity-Enhanced 15N Static NMR in Solids by 1H Indirect Detection”, J. Magn. Reson. 150, 43-48 (2001).
1999 – 2000
25
D. Huster, S. Yamaguchi, and M. Hong, “Efficient b-Sheet Identification in Proteins by Solid-State NMR spectroscopy”, J. Am. Chem. Soc. 122, 11320-11327 (2000).
24
M. Hong, “Solid-State NMR Determination of 13Cα Chemical Shift Anisotropy for the Identification of Protein Secondary Structure”, J. Am. Chem. Soc. 122, 3762-3770 (2000).
23
C.M. Rienstra, M. Hohwy, M. Hong and R.G. Griffin, “2D and 3D 15N-13C-13C NMR chemical shift correlation spectroscopy of solids: assignment of MAS spectra of peptides”, J. Am. Chem. Soc. 122, 10979-10990 (2000).
22
D.J. Harris, T.J. Bonagamba, M. Hong and K. Schmidt-Rohr, “Conformation of Poly(ethylene oxide)-Hydroxybenzene Molecular Complexes Studied by Solid-state NMR”, Macromolecules 33, 3375-3381 (2000).
21
E.R. deAzevedo, S.B. Kennedy and M. Hong, “Determination of Slow Motions in Extensively Isotopically Labeled Proteins by Magic-Angle-Spinning 13C-Detected 15N Exchange NMR”, Chem. Phys. Lett. 321, 43-48 (2000).
20
M. Hong, “Resonance Assignment of 13C, 15N Labeled Proteins by Two- and Three-Dimensional Magic-Angle-Spinning NMR”, J. Biomol. NMR 15, 1-14 (1999).
19
M. Hong and K. Jakes, “Selective and Extensive 13C Labeling of a Membrane Protein for Solid-State NMR Investigation”, J. Biomol. NMR 14, 71-74 (1999).
18
M. Hong, “Determination of Multiple f Torsion Angles in Solid Proteins by Selective and Extensive 13C Labeling and Two-Dimensional Solid-State NMR”, J. Magn. Reson. 139, 389-401 (1999).
17
D. Sandström, M. Hong and K. Schmidt-Rohr, “Identification and Mobility of Deuterated Sites in Peptides and Proteins by 2H-13C Correlation in Solid-State NMR”, Chem. Phys. Lett. 300, 213-220 (1999).
16
M. Hong, “Solid-State Dipolar INADEQUATE NMR Spectroscopy with a Large Double-Quantum Spectral Width”, J. Magn. Reson. 136, 86-91 (1999).
1997 – 1998
15
A.T. Petkova, M. Baldus, M. Belenky, M. Hong, R.G. Griffin, J. Herzfeld, “Backbone and side chain assignment strategies for multiply labeled membrane peptides and proteins in the solid state”, J Magn. Reson. 160, 1-12 (2003).
14
M. Hong, J.D. Gross, W. Hu and R.G. Griffin, “Determination of the Peptide Torsion Angle f by 15N Chemical Shift and 13Cα–1Hα Dipolar Tensor Correlation in Solid-State MAS NMR“, J. Magn. Reson. 135, 169-177 (1998).
13
M. Hong and R.G. Griffin, “Resonance Assignments for Solid Peptides by Dipolar-Mediated 13C/15N Correlation Solid-State NMR”, J. Am. Chem. Soc. 120, 7113-7114 (1998).
12
P.R. Costa, J.D. Gross, M. Hong and R.G. Griffin, “A NCCN 2Q-HLF Experiment for Y Torsion Angle Measurements in Peptides”, Chem. Phys. Lett. 280, 95-103 (1997).
11
M. Hong, J.D. Gross, C.M. Rienstra, R.G. Griffin, K.K. Kumashiro and K. Schmidt-Rohr, “Coupling Amplification in 2D MAS NMR and its Application to Torsion Angle Determination in Peptides”, J. Magn. Reson. 129, 85-92 (1997).
10
M. Hong, J.D. Gross and R.G. Griffin, “Site-resolved Determination of Peptide Torsion Angle Phi from the Relative Orientations of Backbone N-H and C-H Bonds by Solid-State NMR”, J. Phys. Chem. 101, 5869-5874 (1997).
1994 – 1996
9
S. Caldarelli, M. Hong, L. Emsley and A. Pines, “Measurement of Carbon-Proton Dipolar Coupling in Liquid Crystals by Local Dipolar Field NMR Spectroscopy”, J. Phys. Chem. 100, 18696-18701 (1996).
8
M. Hong, A. Pines and S. Caldarelli, “Measurement and Assignment of Long-Range C-H Dipolar Couplings in Liquid Crystals by Two-Dimensional NMR Spectroscopy”, J. Phys. Chem. 100, 14815-14822 (1996).
7
M. Hong, K. Schmidt-Rohr and Zimmerman H, “Conformational Constraints on the Headgroup and sn-2 Chain of Bilayer DMPC from NMR Dipolar Couplings”, Biochemistry 35, 8335-8341 (1996).
6
K. Schmidt-Rohr and M. Hong, “Information on Bond Orientation Distributions in Lipids and Liquid Crystals from Segmental Order Parameters”, J. Phys. Chem. 100, 3861-3866 (1996).
5
M. Hong, K. Schmidt-Rohr and D. Nanz, “Study of Phospholipid Structure by 1H, 13C, and 31P Dipolar Couplings from Two-Dimensional NMR”, Biophys. J. 69, 1939-1950 (1995).
4
M. Hong and K. Schmidt-Rohr, “DISTINCT NMR for Sign Determination of C-H Dipolar Couplings in Liquid-Crystalline Lipids”, J. Magn. Reson. B 109, 284-290 (1995).
3
M. Hong, K. Schmidt-Rohr and A. Pines, “NMR Measurement of Signs and Sizes of C-H Dipolar Couplings in Lecithin”, J. Am. Chem. Soc. 117, 3310-3311 (1995).
2
D. Nanz, M. Ernst, M. Hong, M.A. Ziegeweid, K. Schmidt-Rohr and A. Pines, “Low-Power Decoupling with Windowless Multiple-Pulse Sequences for High-Resolution Spectra of Orientationally Ordered Samples”, J. Magn. Reson. A 113,169-176 (1995).
1
Y.K. Lee, L. Emsley, R.G. Larsen, K. Schmidt-Rohr, M. Hong, L. Frydman, G.C. Chingas and A. Pines, “Three-Dimensional Variable-Angle Nuclear Magnetic Resonance Exchange Spectroscopy without Rotor Axis Hopping”, J. Chem. Phys. 101 (3), 1852-1864 (1994).
$ denotes equal contribution